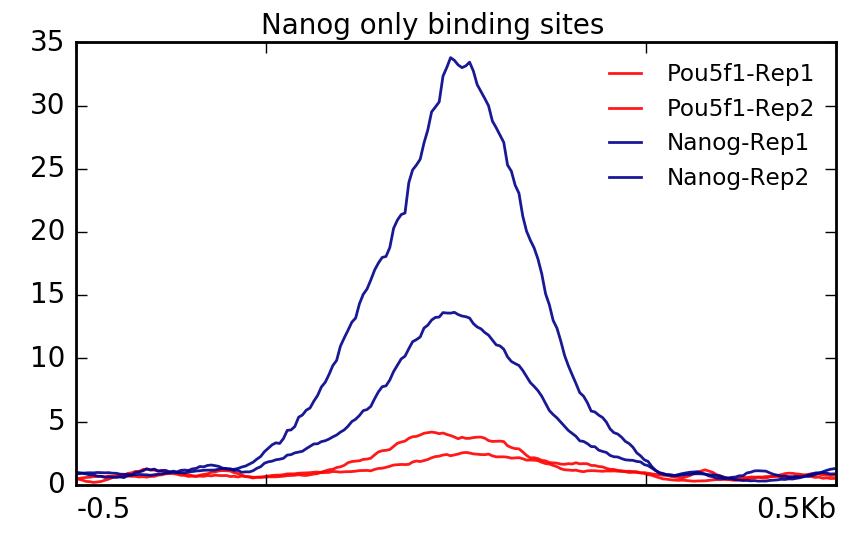
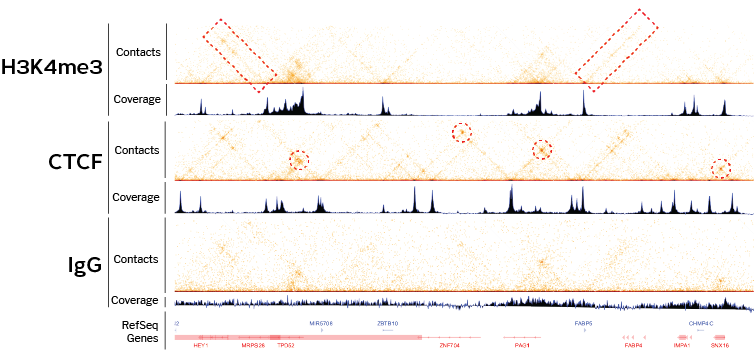
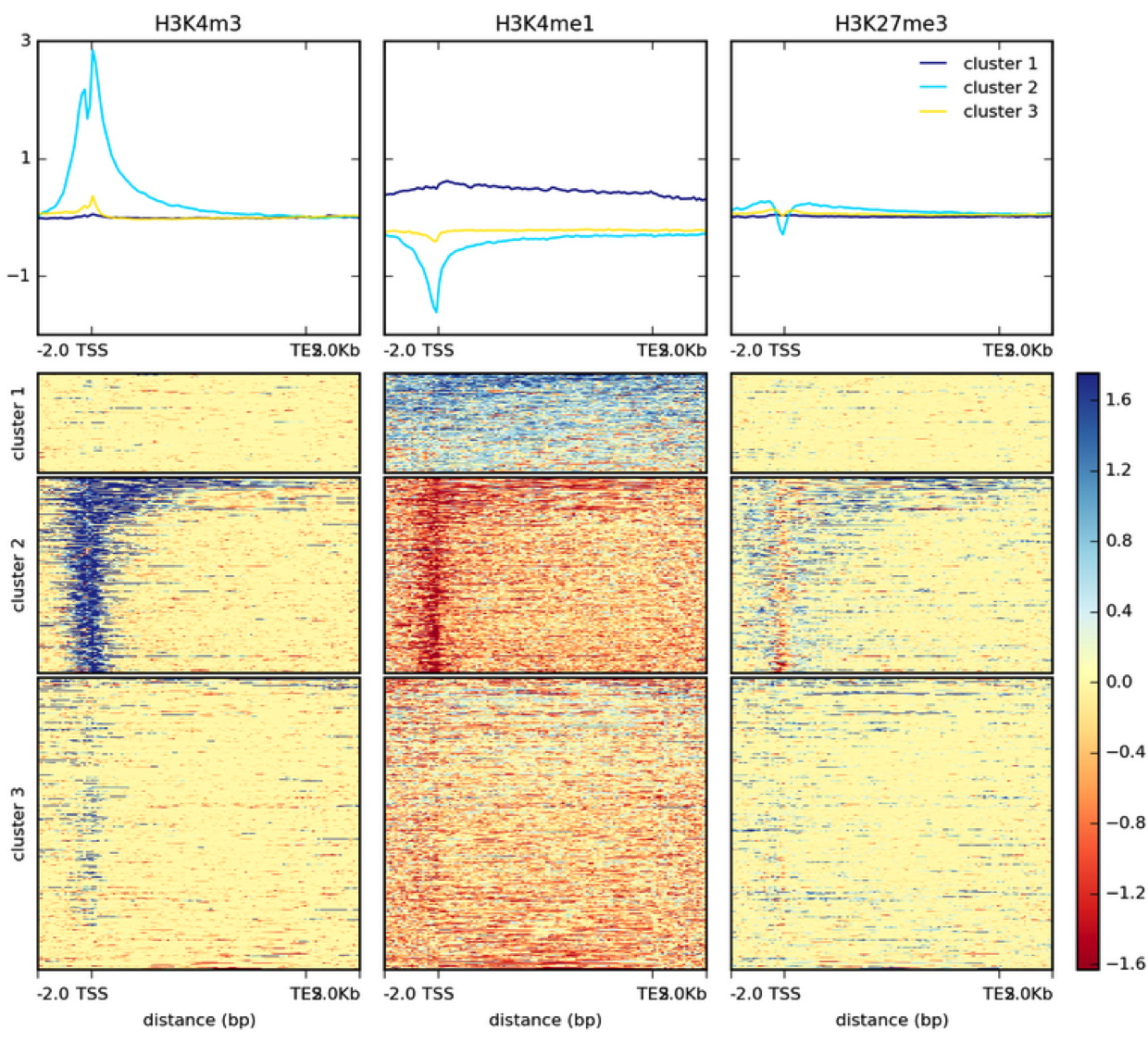
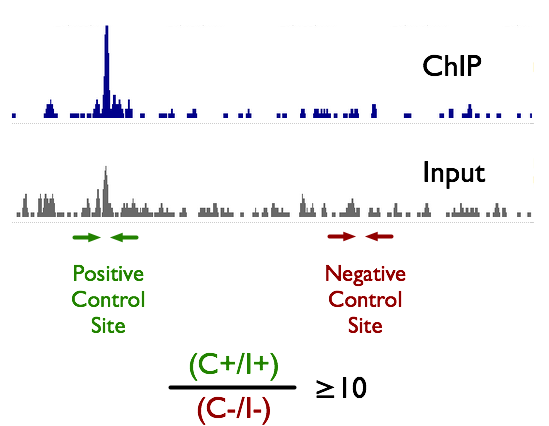
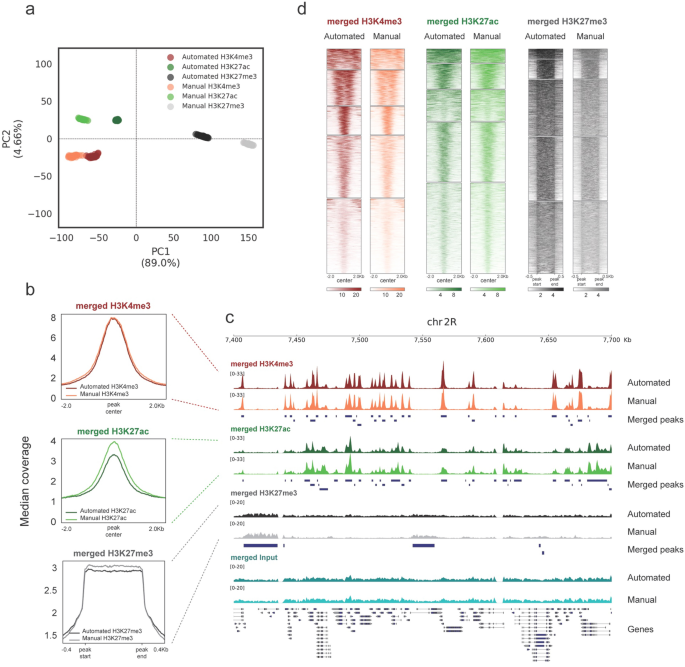
ChIP-Seq deconvolution maps are highly reproducible. (A) Comparison of... | Download Scientific Diagram

Seq-ing answers: Current data integration approaches to uncover mechanisms of transcriptional regulation - ScienceDirect

Sequence coverage dominates the raw ChIP-Seq profiles for UBF and RPI.... | Download Scientific Diagram
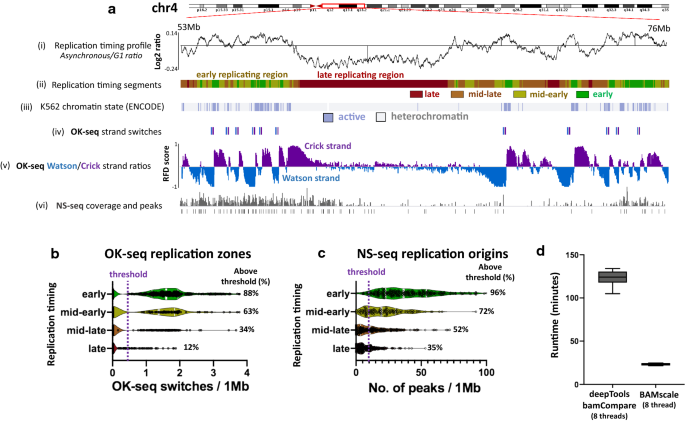
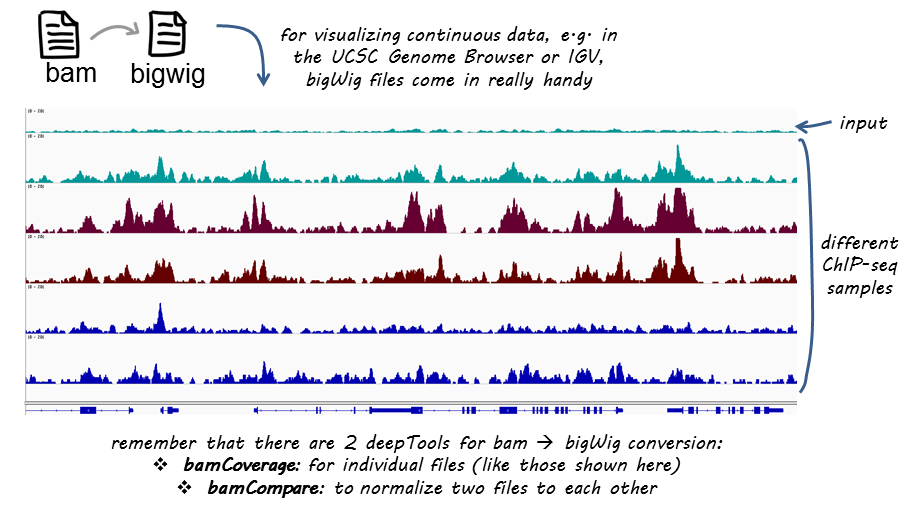
BAMscale: quantification of next-generation sequencing peaks and generation of scaled coverage tracks | Epigenetics & Chromatin | Full Text
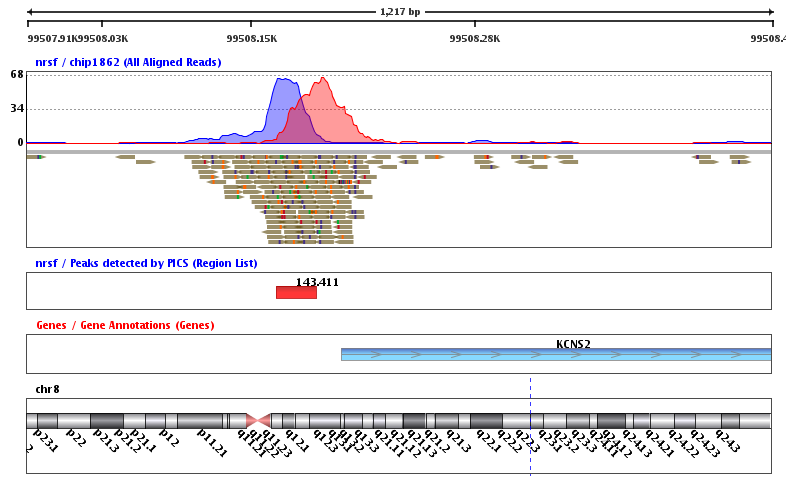
CNN-Peaks: ChIP-Seq peak detection pipeline using convolutional neural networks that imitate human visual inspection | Scientific Reports

Vezf1 protein binding sites genome-wide are associated with pausing of elongating RNA polymerase II | PNAS

TAF-ChIP: an ultra-low input approach for genome-wide chromatin immunoprecipitation assay | Life Science Alliance